The importance of data integration
Cohort studies produce invaluable data typically including a range of different data types, such as clinical (e.g. patient health records, blood markers, treatment details) as well as multi-omics data (such as genomic sequence, transcriptomic or proteomic data). However, often these data types are siloed in archives specific for each type. Data integration, i.e, linking these data types on a participant level across different repositories, adds more depth to the dataset by bringing the pathogen and host data into context, allowing a more comprehensive analysis of the entire collection of data.
Here we describe the first example of linking and sharing a multi-omics SARS-CoV-2 cohort dataset with multiple time points, via the European Molecular Biology Laboratory-European Bioinformatics Institute’s (EMBL-EBI) infrastructure.
Who is the showcase intended for?
This showcase is intended for any group who would like to FAIRly share cohort study data, and would like to better understand how this can be performed. More broadly, the linking mechanism described below is applicable to any group whose research involves different types of molecular data being obtained from the same biological sample. The showcase will also be useful for those wishing to access linked clinical and multi-omics data from cohort studies.
Sharing the linked EMC Pilot Cohort Dataset
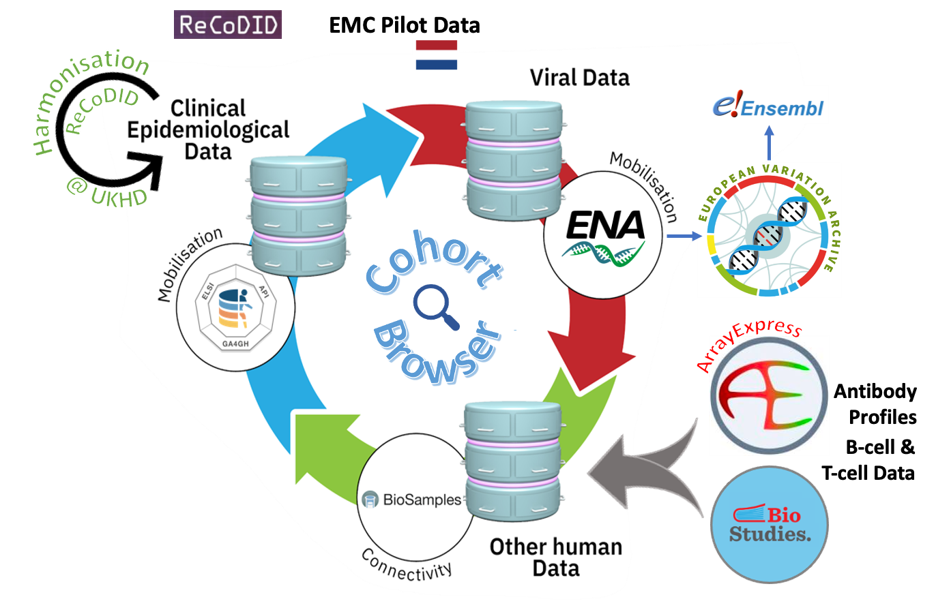
Figure 1. An overview of the data integration process across EMBL-EBI archives. At the ENA, all SARS-CoV-2 sequence data undergo in-house systematic analysis, and these products feed into the European Variation Archive (EVA) and Ensembl. More details can be found from the pre-print here.
The team at Erasmus Medical Centre (EMC Rotterdam, NL) collected data from a cohort of 151 Polymerase Chain Reaction (PCR)-confirmed COVID-19 individuals who had been admitted to the hospital with a respiratory infection or respiratory failure in 2020-2021. After harmonisation of the clinical-epidemiological data using the International Severe Acute Respiratory and emerging Infection Consortium’s (ISARIC) data dictionary for the ISARIC COVID-19 Case Report Form (carried out by ReCoDID partners at the university hospital in Heidelberg, Germany), the sensitive, pseudonymised clinical-epidemiological data were submitted to the European Genome-phenome Archive (EGA), SARS-CoV-2 sequences were deposited in European Nucleotide Archive (ENA) and ArrayExpress/BioStudies was used to archive antibody profiles as well as B-cell and T-cell data (see Figure 1).
The various data types were linked on participant level by utilising the BioSamples data archive which can capture relationships between samples. In a hierarchical structure a top-level sample ID was created which represents the patient (and the associated EGA record). Further BioSample IDs were created for each time point and/or data type and these were linked to the top-level sample ID of this patient, creating a link between data types on participant level (see Figure 2).
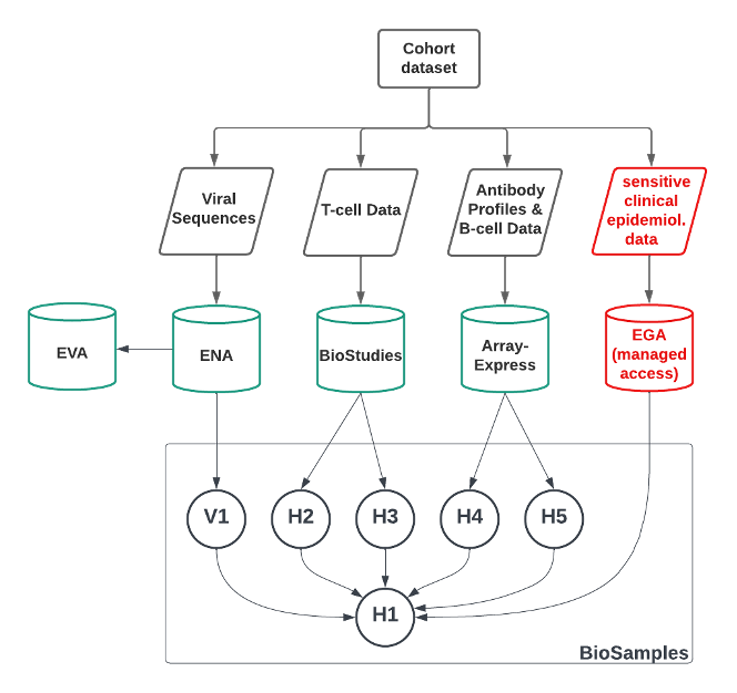
Figure 2. Schematic of participant level linking of Biosamples. ‘V’ refers to Viral, while ‘H’ refers to non-sensitive human samples (e.g. representing data types, or the top-level COVID-19 patient).
Composition of the EMC Pilot Cohort Dataset:
The composition of the EMC SARS-CoV-2 Dataset is summarised below and in Figure 3.
- Clinical-epidemiological information is available for all 151 patients that tested PCR-positive for SARS-CoV-2. For 77 of those, additional data types are available and these have also been submitted and linked; for the remaining 74 patients only clinical-epidemiological data is available. (Archived in European Genome-phenome Archive (EGA), controlled access managed through a Data Access Committee (DAC));
- 80 SARS-CoV-2 sequences are available for 63 patients, with 1-4 sequences per patient. (Archived in European Nucleotide Archive (ENA), open access);
- Antibody profiles are available for 40 patients with 2-5 time points per patient, with a total of 147 data points. (Archived in ArrayExpress/BioStudies, open access);
- T-cell data are available for 28 patients with 1, 3 or 4 time points per patient and a total of 51 data points. (Archived in ArrayExpress/BioStudies, open access);
- B-cell data are available for 17 patients with either one or 3 time points and a total of 29 data points. (Archived in ArrayExpress/BioStudies, open access).
All sample records for this dataset, together with the appropriate links, can be viewed in the BioSamples database here.
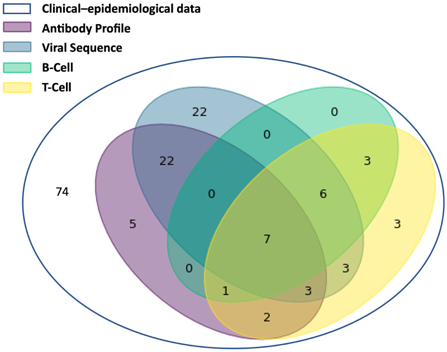
Figure 3. A Venn diagram showing the different combinations of data types for all 151 patients in the EMC study.
The Cohort Browser - improving the Findability of Cohort Datasets
While the BioSamples database is key to capturing the linking of data types on a participant level, the Pathogens Portal Cohort Browser presents a range of study-level information about each cohort. Similar to a shop window, it enhances the findability of the datasets and as an integral part of the Pathogens Portal, serves as the primary entry point into accessing cohort data.
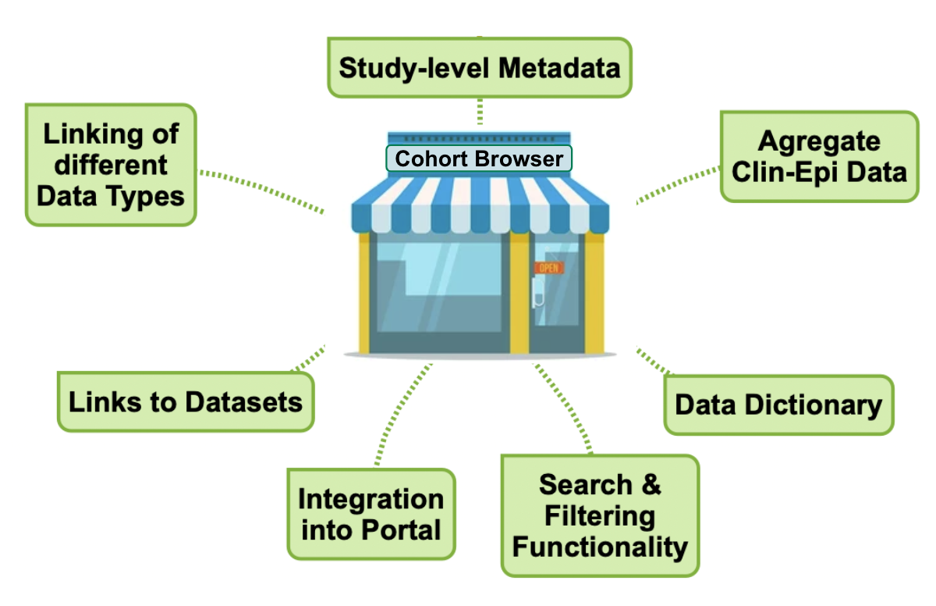
Figure 4. The cohort browser brings together study-level information, links to the datasets and provides search and filtering functionality to improve data discoverability.
The Cohort Browser lists discovery metadata of cohort studies and provides links to the available data types. Where possible, basic aggregate data are also included and future search and filtering functionality will allow users to locate datasets of interest.
What can you use the showcase for?
- this showcase provides a proof of concept for linking and sharing of entire cohort datasets in line with FAIR principles, with data sharing being as open as possible and as closed as necessary.
- this showcase provides the first public, cohort study example of linking different biological data types, at the sample-level, across dedicated archives.
- the applied linking process significantly increases the interoperability between the bespoke archives for the various data types, making it easier to access and analyse them.
- linking these data types on a participant level also adds more depth to the dataset by bringing the pathogen and host data into context, allowing a more comprehensive analysis. This is particularly important in public health but will equally provide a better understanding of infectious diseases in the context of both pathogen & their host factors.
Acknowledgements
ReCoDID Project Team Members
The work presented here was a coordinated effort of multiple teams covering the different aspects of the process:
Data collection and submissions were coordinated by the Viroscience department at Erasmus Medical Centre (EMC), Rotterdam, NL:
Clara Amid, Janko van Beek, Gijs van Nierop, Rory de Vries, Marion Koopmans.
Harmonisation of clinical-epidemiological data was performed at the University Hospital Heidelberg, Germany:
Frank Tobian, Thomas Jänisch.
The infrastructure for data sharing and support for the data linking process was provided by EMBL-EBI, Hinxton, UK:
Gabriele Rinck, Zahra Waheed, Nadim Rahman, Marcos Casado Barbero, Mallory Freeberg, Dipayan Gupta, Anja Füllgrabe, Ugis Sarkans, Guy Cochrane.
Additional people who have contributed:
Coline Thomas, Marianna Ventouratou, Suran Jayathilaka and Vishnukumar Balavenkataraman Kadhirvelu.
Support
ReCoDID project is funded by the European Union’s Horizon 2020 Research and Innovation Programme, grant agreement N° 825746, and the Canadian Institutes of Health Research, Institute of Genetics (CIHR-IG), grant agreement N° 01886-000.
More information
Skip tool tableTools and resources on this page
| Tool or resource | Description | Related pages | Registry |
|---|---|---|---|
| ArrayExpress | ArrayExpress is a database of functional genomics experiments that can be queried and the data downloaded. It includes gene expression data from microarray and high throughput sequencing studies. Data is collected to MIAME and MINSEQE standards. Experiments are submitted directly to ArrayExpress or are imported from the NCBI GEO database. | Human biomolecular data | Tool info Standards/Databases Training |
| BioSamples | BioSamples stores and supplies descriptions and metadata about biological samples used in research and development by academia and industry. Samples are either 'reference' samples (e.g. from 1000 Genomes, HipSci, FAANG) or have been used in an assay database such as the European Nucleotide Archive (ENA) or ArrayExpress. It provides links to assays and specific samples, and accepts direct submissions of sample information. | Human biomolecular data | Tool info Standards/Databases Training |
| BioStudies | The BioStudies database holds descriptions of biological studies, links to data from these studies in other databases at EMBL-EBI or outside, as well as data that do not fit in the structured archives at EMBL-EBI. The database can accept a wide range of types of studies described via a simple format. It also enables manuscript authors to submit supplementary information and link to it from the publication. | FAIR data | Tool info Standards/Databases Training |
| European Genome-phenome Archive (EGA) | The European Genome-phenome Archive (EGA) is a service for permanent archiving and sharing of personally identifiable genetic, phenotypic, and clinical data generated for the purposes of biomedical research projects or in the context of research-focused healthcare systems. Access to data must be approved by the specified Data Access Committee (DAC). | FAIR data Human biomolecular data Human clinical and hea... | Tool info Standards/Databases Training |
| European Nucleotide Archive (ENA) | Provides a record of the nucleotide sequencing information. It includes raw sequencing data, sequence assembly information and functional annotation. | Pathogen characterisation Human clinical and hea... Pathogen characterisation Human biomolecular data An automated SARS-CoV-... Using the ENA data sub... SARS-CoV-2 sequencing ... | Tool info Standards/Databases Training |
| ISARIC COVID-19 Case Report Form | The ISARIC-WHO Case Report Forms (CRFs) should be used to collect data on individuals presenting with suspected or confirmed COVID-19, with the aim to standardise clinical data to improve patient care and inform the public health response. | Standards/Databases | |
| Pathogens Portal | The Pathogens Portal, launched in July 2023, is an invaluable resource for researchers, clinicians, and policymakers who need access to the latest and most comprehensive datasets on pathogens. The portal is a collaborative effort between the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) and partners. | Pathogen characterisation The Swedish Pathogens ... | Standards/Databases Training |
| Pathogens Portal Cohort Browser | The Pathogens Portal Cohort Browser presents discovery metadata of infectious disease cohort datasets and provides links to the associated datasets within ELIXIR Core Data Resources; search and filtering functionalities enable users to identify cohort studies of interest in a convenient manner. |


