Introduction
Sequencing initiatives in Estonia began in Spring 2020. The KoroGeno-EST-3 and KoroGeno-EST-2022 projects took over the sequencing activities in May 2021, and their work concluded at the end of 2022. Here, we describe the workflow of samples and data employed by the KoroGeno-EST-3 and KoroGeno-EST-2022 projects .
Who is the KoroGeno-Est showcase intended for?
This showcase provides a high-level overview of the full workflow that was used to monitor the SARS-CoV-2 situation in Estonia by sequencing a proportion of positive PCR samples in the country. The showcase can be beneficial for a general overview but also trigger contact with research groups or health boards that need to set up their automatic data handling and sequencing pipelines.
What is the showcase?
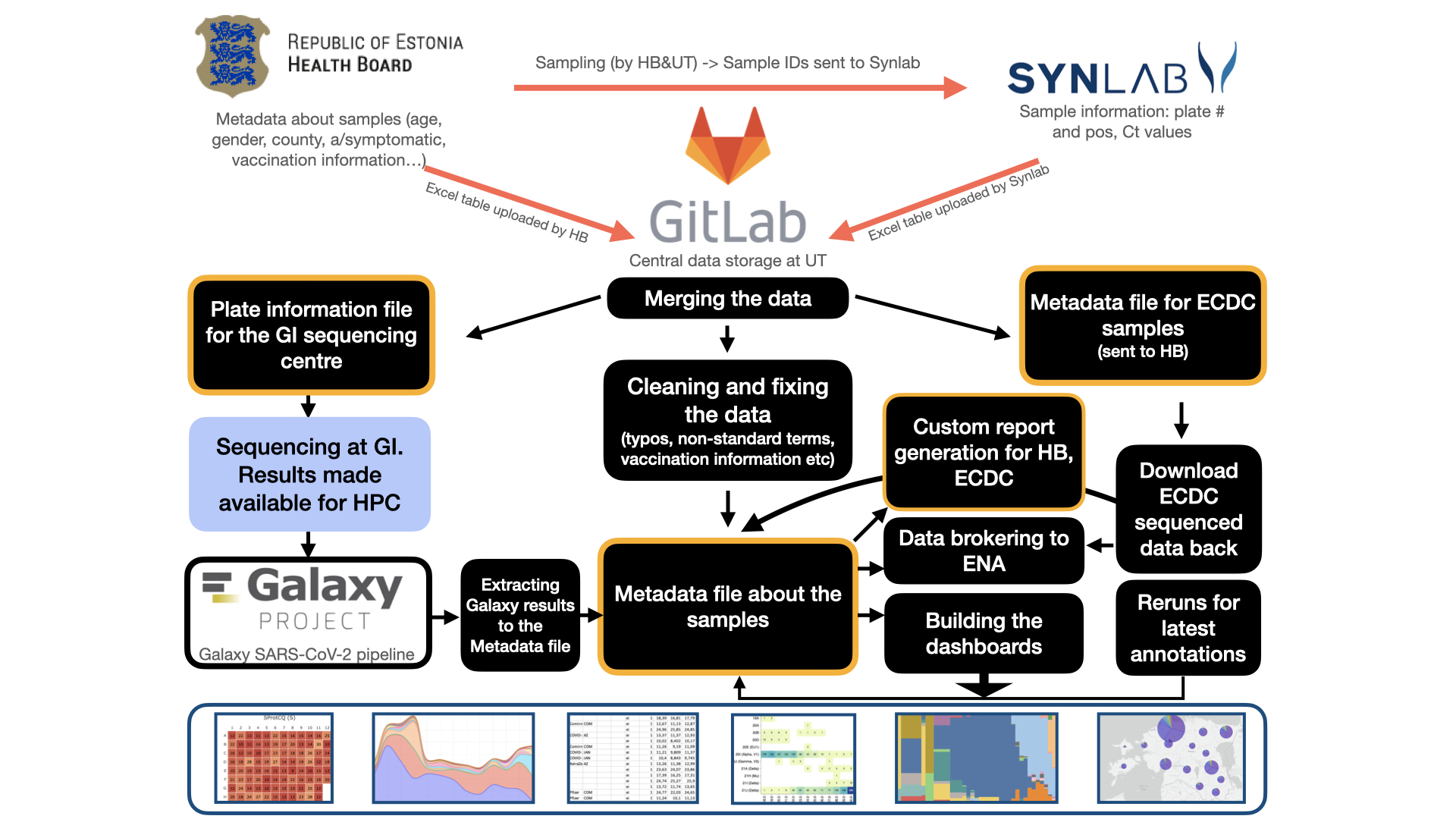
The figure illustrates major analysis steps carried out in Estonia to sequence and analyse SARS-CoV-2 PCR positive samples
In Estonia the SARS-CoV-2 sequencing was carried out in collaboration with SYNLAB Eesti, the Republic of Estonia Health Board and the University of Tartu. SYNLAB Eesti provided information on the basic sample metadata and PCR outcome, the Health Board provided additional sample metadata on e.g. age, gender, county, vaccination information.
The central data storage was at a GitLab instance at the University of Tartu. The University of Tartu research groups merged the data, created 96-well plate layout files for sequencing, collected the sequencing results, generated custom reports, interactive dashboards and brokered data to European Nucleotide Archive (ENA). In addition to data sequenced in Estonia, also Estonian samples sequenced at ECDC were downloaded from there and analysed in a similar manner to provide an accurate overview of the situation.
The aim of KoroGeno-EST
The aim of KoroGeno-EST-3 and KoroGeno-EST-2022 (hereafter, Study) studies was to sequence and conduct molecular-epidemiology analysis for more than 15 000 SARS-CoV-2 full genomes from 1 May to 31 December 2022 in Estonia. The number of weekly sequenced strains depended directly on the epidemiological situation, including the number of newly diagnosed SARS-CoV-2 cases, as well as the need to monitor clinically, demographically, or mutationally significant strains.
Based on the sequenced samples, weekly reports were prepared for the Republic of Estonia Health Board (hereafter, Health Board (HB)) and “European Centre for Disease Prevention and Control (ECDC)”. In collaboration with the HB, SARS-CoV-2 genomic data was used to analyse infection clusters to trace newly infected subjects.
In addition, the study identified the prevalence of variants associated with higher pathogenicity or infectiousness or corresponding mutations. Based on these data, authorities got a better overview of circulating SARS-CoV-2 strains as well as cases from abroad. In general, the current study made a significant contribution to the governmental decision-making process and helped more efficiently suppress the spread of the COVID-19 epidemic.
Metadata and sample acquisition
Throughout most of the pandemic, SYNLAB Eesti testing sites in Estonia provided free SARS-CoV-2 PCR testing for all individuals living in the country. Free testing was discontinued for general population in May 2022 and for risk groups in November 2022. Self-paid testing has been available since 2020.
Sample metadata
Initially, at the beginning of the pandemic, all persons testing positive were contacted by the Health Board authorities and asked to provide details of their recent travels, family and work situation, in addition to more standard metadata of age, gender and place of living. This type of data collection was ongoing till spring 2022, after which only more general metadata was collected (gender, age, county). Once vaccination was made available to the public in spring 2021, also vaccination information was first self-reported, and later data including information on hospitalisation was systematically obtained from the Health and Welfare Information Systems Centre (TEHIK). In addition hospitalisation information was provided by the Health Board who obtained it from TEHIK.
PCR testing
The laboratory of SYNLAB Eesti carried out PCR testing. They captured Ct values and particular mutation sites for all positively tested samples. The social security codes that uniquely identify a person in Estonia were shared with the Health Board. Health Board together with the virology experts from the University of Tartu chose samples for sequencing based on various parameters.
In general, the samples were carefully selected to ensure representation of the Estonian demographic and geographic subgroups, with additional samples taken from outbreaks that occurred at hospitals, elderly homes or work places. The data at individual level was linked at the Health Board and sample IDs from SYNLAB Eesti lab and the matching metadata IDs were created. Both the Health Board and SYNLAB Eesti uploaded the data about the samples to an access restricted GitLab repository. The University of Tartu team received only pseudonymised data.
In addition to sequencing carried out in Estonia, samples were also sent to ECDC. The metadata for these samples followed the same pipeline as above. But the resulting sequences were downloaded from dedicated ECDC websites and bioinformatically processed in the same manner as the samples sequenced in Estonia.
Metadata cleaning and standardisation
The metadata was semi-automatically cleaned and automatically standardised by custom-developed Python scripts. Whenever available, the following metadata fields were cleaned and standardised - age, age group, county, region of country, symptom onset date, place of infection, vaccine producer and dose.
Sequencing and sequence analysis
Since May 2021 the samples were transported from SYNLAB Eesti and hospitals laboratories directly to the University of Tartu Genome Centre Core lab who carried out the sequencing using Illumina NextSeq sequencer. Based on the sample information and original location on the laboratory plates the automatic pipeline generated the 96-well plate layouts needed to carry out the sequencing.
The sequencing was carried out using Arctic3 and later Arctic4 libraries. At times also NebNext Arctic protocols were tested out.
The raw sequences were initially analysed by the Illumina Dragen pipeline at the University of Tartu Genome Centre Core lab, then in parallel analysed with the Galaxy workflow developed by the Galaxy community led by Wolfgang Maier and eventually only analysed by the Galaxy workflow. The pipeline includes removing the human reads from the raw sequences, mapping the reads to the consensus sequence of SARS-CoV-2, identifying mutations, creating multiple alignments, calling the Pangolin lineages and NextClade clades.
The work was carried out in the Galaxy University of Tartu hosted by the ETAIS at the University of Tartu HPC. All the identified information on the samples was automatically fetched from the dedicated Galaxy address to the GitLab repository and merged with the rest of the metadata about the samples.
Generating custom reports
Based on the templates provided by the ECDC, reports were filled in automatically based on the sequencing results. These reports included variants of concern (VoCs), specific set of mutations or deletions defined by the ECDC that changed over time. Similar reports were also generated for the Health Board to inform the Government.
Dashboards
Description of the closed dashboard
The data table of all the samples was made available at the access restricted website for the Health Board employees and project participants to provide epidemiological and virological consultations.
The database included all metadata provided for the samples, sequencing outcomes and was interactive providing filtering and sorting among other customisation options. In addition data could be downloaded for further custom analysis. Also, the closed website of the project dashboard included different interactive plots that allowed the users to pick the variables to be plotted (e.g. Ct values against vaccination status, county against lineages), and also lineage overview where individual lineages could be included or dismissed, and time frame could be selected.
To carry out visual inspections as part of the quality control of the sequencing outcomes, mutation and Ct value plots were included for each sequenced plate. In all the dashboards, users could select if they wanted to include data only sequenced in Estonia, by ECDC or both.
Public dashboards
In addition to the access restricted dashboards, a regularly updated Estonian COVID-19 Data Portal public dashboard was also deployed. It gave an overview of clade distribution across counties, regions and age groups, alongside the percentage of all positive samples that have been sequenced in Estonia. In order to illustrate the virus evolution a public Nextstrain Auspice instance was set up. In regular time intervals, all existing samples were rerun to obtain the latest lineage annotations.
Data brokering to ENA
To upload data to the European Nucleotide Archive (ENA) we registered our research projects at the ENA following the guidelines. At the beginning, individual projects were created for each data batch. Later on, multiple batches of data were joined under the same project. The data broker manually generated ENA metadata files following the ENA virus pathogen reporting standard checklist ERC000033, and using the equivalent spreadsheet metadata template. The preparatory work for data brokering to ENA was done by a dedicated Galaxy workflow. A typical workflow from the extraction of Galaxy results data and metadata submission took about 1 hour per plate (94 wells).
Estonia used the Galaxy ENA upload Galaxy tool that is described in The Galaxy Toolshed. The tool makes use of the ENA upload CLI. Documentation is available in dedicated pages in the RDM Guide, and a standalone publication.
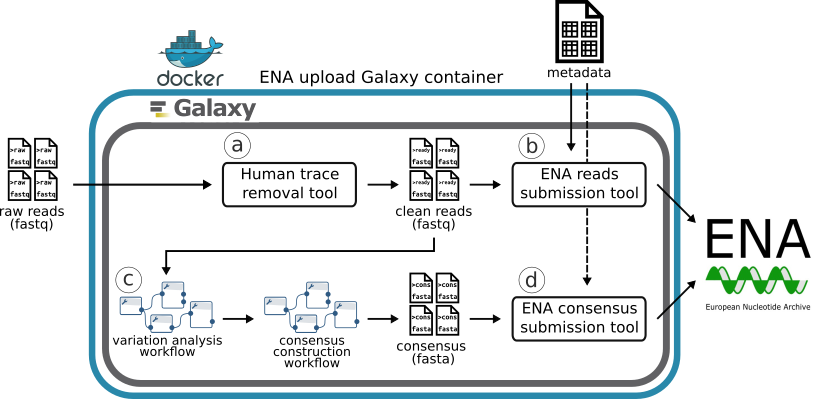
An overview of SARS-CoV-2 raw reads and metadata upload steps (a-d) to ENA enabled by Galaxy. Adapted from A SARS-CoV-2 sequence submission tool for the European Nucleotide Archive publication
All the sequenced raw reads results were uploaded to the University of Tartu Galaxy instance. From there the data manager selected a batch and successfully sequenced samples into a working collection. Forward and reverse sequences were paired up and run through the workflow for removal of traces of human genetic information from the raw sequence reads (uses Metagen-FastQC).
In Galaxy ENA uploader metadata checklist, ERC000033, was marked and the filled-out template was uploaded. The runs input format was selected from a paired collection (that was human sequence free), an affiliation centre was added, and the ENA upload tool was run.
As of March 2023, over 23 000 Estonian SARS-CoV-2 sequences have been uploaded to ENA. The majority were sequenced during the KoroGeno-EST studies (14251), complemented by 9978 successfully sequenced samples from ECDC.
Considerations in Estonia
- Estonia has a single national Health Board
- e-Estonia and the X-road (e-Identity, e-Health Records, e-Governance) infrastructure allows easy data linking across data sources using the personal identification number.
- There has been previous studies and cooperation between the Health Board and the University of Tartu in virus monitoring (HIV, Flu)
- There were no additional local laws restraining the study
- While the code of the dashboards and data handling is not publicly available, the team is happy to consult researchers facing similar challenges. The best way is to send an email to ELIXIR-Estonia. Estonia adapted and used the original publicly available Galaxy workflows COVID-19: consensus construction, COVID-19: variation analysis on ARTIC PE data, COVID-19: variation analysis reporting.
What can you use the SHOWCASE for?
We provided the above information on how we set up an automatic pipeline of handling SARS-COV-2 sample sequencing in Estonia during the COVID-19 pandemic in 2021-2022. The pipeline was successfully used to sequence thousands of samples in Estonia, combine the data from ECDC sequencing initiative, and provide tools for the Health Board and weekly reports nationally and internationally. The team is happy to share further insights, code snippets and visualisation information based on requests.
External links to resources relevant to the KoroGeno-Est showcase
- Estonian Covid19Dataportal
- Overview of the SARS-CoV-2 projects (in Estonian) - Information on how to register a study to ENA
- ENA virus pathogen reporting standard checklist
- ENA metadata templates
- Galaxy wrapper for the ENA upload tool
- Submission of SARS-Cov-2 raw reads to ENA in Galaxy
- General Guide On ENA Data Submission by ENA
- European Nucleotide Archive Quick tour by EMBL-EBI
- Example metadata template ERC000033
Acknowledgements
The study was approved by Research Ethics Committee of the University of Tartu.
Team members of KoroGeno-Est
Core team at University of Tartu: Virology and epidemiology: Kristi Huik (main coordinator of the project); Aare Abroi, Irja Lutsar, Radko Avi, Merit Pauskar, Taavi Päll, Arina Shablinskaja, Kai Truusalu, Dagmar Hoidmets, Ene-Ly Jõgeda, Eveli Kallas, Katrin Kaarna (project management) Sequencing: Tuuli Reisberg (coordinator of sequencing), Mait Metspalu, Lili Milani, Steven Smit Bioinformatics: Hedi Peterson (coordinator of bioinformatic), Ulvi Gerst Talas, Ivar Koppel, Ott Eric Oopkaup, Erik Jaaniso, Ivan Kuzmin, Uku Raudvere; Heleri Inno, Diana Pilvar (data managers)
Synlab team: Andrio Lahesaare, Paul Naaber, Krislin Raus, Kaspar Ratnik
Estonian Health Board team: Mari-Anne Härma, Heiki Niglas, Liidia Dotsenko, Olga Sadikova, Hanna Sepp, Heleene Suija, Jevgenia Epštein
Support
The KoroGeno-EST studies were conducted in cooperation with the University of Tartu, the Estonian Health Board and SYNLAB Eesti OÜ during 2021-2022. The KoroGeno-EST-3 and KoroGeno-EST-2022 projects were financed by the Republic of Estonia’s Ministry of Education and Research.
Related pages
More information
Training
Skip tool tableTools and resources on this page
| Tool or resource | Description | Related pages | Registry |
|---|---|---|---|
| ENA upload CLI | Command line tool (CLI) allowing easy submission of data and respective metadata to the European Nucleotide Archive (ENA) using tabular files or an excel spreadsheet. The tool allows programatic submission of all ENA objects (study, sample, run and experiment) without the need of logging in to the Webin interface. This also includes client side validation using ENA checklists and releasing the ENA objects. | Using the ENA data sub... | |
| ENA upload Galaxy tool | Galaxy tool wrapper of the ENA upload CLI to submit experimental data and respective metadata to the European Nucleotide Archive (ENA). | Using the ENA data sub... | |
| Estonian COVID-19 Data Portal | Estonian instance of the COVID-19 Data Portal. Among other information, served Estonian SARS-CoV-2 sequencing dashboards. | The Swedish Pathogens ... | |
| European Nucleotide Archive (ENA) | Provides a record of the nucleotide sequencing information. It includes raw sequencing data, sequence assembly information and functional annotation. | Pathogen characterisation Human clinical and hea... Pathogen characterisation Human biomolecular data An automated SARS-CoV-... Using the ENA data sub... Linked pathogen and ho... | Tool info Standards/Databases Training |
| Galaxy University of Tartu | The University of Tartu Galaxy instance. Enables local university users to run their analyses in the Galaxy environment. Was heavily used during the KoroGenoEST sequencing studies. | ||
| Metagen-FastQC | Cleans metagenomic reads to remove adapters, low-quality bases and host (e.g. human) contamination. | Using the ENA data sub... | |
| Nextstrain Auspice | Estonian local instance of the Nextstrain Auspice application that serves SARS-CoV-2 phylogenetic data |


